Virtual liver model reduces risk of overdose
A virtual model of the human liver has been developed by researchers at Indiana University’s Biocomplexity Institute to better understand how the organ metabolizes acetaminophen, a common non-prescription painkiller and fever-reducer used in over-the-counter drugs such as Tylenol.
The study, reported in the peer-reviewed online journal PLOS ONE, suggests that virtual tissues models could play an important role in modern pharmacokinetics, the branch of pharmacology concerned with the movement of drugs within the body.
Specifically, the study employs “virtual tissue” technology developed at the institute to model the distribution of drugs in the human body at multiple scales – time and location in the body – an ability that could help contribute to more personalized prescription guidelines that reduce the risk of overdose.
Acetaminophen metabolism is significant because poisoning from a liver metabolite of acetaminophen is the leading cause of acute liver failure in the United States, resulting in over 33,000 hospital admissions and 500 deaths each year. Acute liver failure can lead to death since the liver is the primary organ responsible for the metabolism and clearance of toxins, including drugs, from the body.
“Although this model of acetaminophen toxicity isn’t currently directed at developing interventions to accidental overdoses, it suggests a surprisingly large degree of difference in sensitivity to the drug across large populations,” said James Sluka, a research scientist at the Biocomplexity Institute, who is a co-lead author on the paper. “According to our model, as many as one in a thousand people may be unusually sensitive to the drug’s toxicity.”
The other co-lead author on the work is Xiao Fu, a graduate student at the Biocomplexity Institute. Other authors are Julio Belmonte, also a graduate student; Sherry Clendenon, a research associate; and Maciej Swat, an associate scientist, all of the Biocomplexity Institute. John Wambaugh, a computational toxicologist with the U.S. Environmental Protection Agency, also contributed to the study.
The senior author on the paper is James A. Glazier, director of the Biocomplexity Institute and a professor in the Department of Intelligent Systems Engineering in the School of Informatics and Computing at IU Bloomington.
The liver acts as a blood flow gateway between the gut and the rest of the body due to the organ’s position between the gastro-intestinal tract and the systematic circulation part of the cardiovascular system. It also plays a key role in preventing nutrient imbalance, which can contribute to high cholesterol, obesity and Type II diabetes.
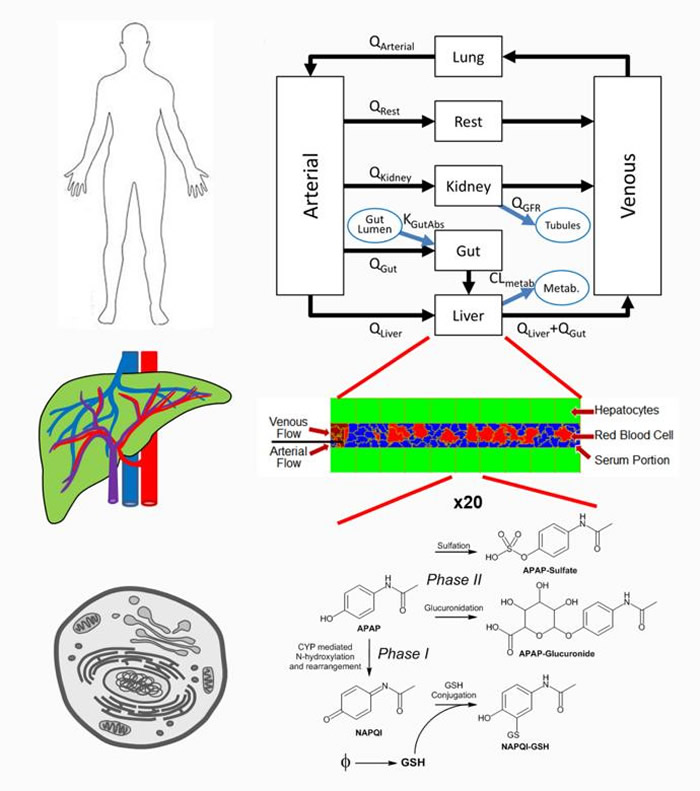
To construct a virtual liver model that took into account “multiple scales” of time and location in the body, Sluka and colleagues used publically available data on acetaminophen distribution and metabolism that reflected the drug’s uptake in the gastrointestinal tract, distribution throughout the body and uptake and metabolism in the liver, and the clearance of the drug and metabolites from the body.
“The lack of significant data in humans beyond models of drug concentration in the blood stream was a major challenge,” Sluka said. “We had to not only depend upon human data but also information from animal- and cell-based studies to create a highly accurate virtual liver.”
At the whole-body level, Sluka and colleagues’ model simulates a drug’s absorption from the gastro-intestinal tract, distribution by the blood to the tissues and organs, and removal by the liver and kidneys.
At the organ level, the model simulates how compounds carried by the blood enter the liver through the hepatic artery and portal vein; flow through the network of sinusoids, which are liver capillary blood vessels; and exit though the organ’s hepatic central vein.
At the subcellular level, the model takes into account how metabolic pathways inside the liver convert chemical compounds into metabolites, which are either transported back into the blood or into the gastrointestinal tract through the bile duct.
Metabolites can also be toxic to liver cells and, in the case of overdose, irreversibly damage the liver. Generally, gene expression patterns within the liver determine which of the organ’s multiple pathway are used to metabolize a chemical.
Using this system, Sluka and colleagues were able to model an oral dose of acetaminophen’s concentration at various points in time in various tissues in the body, especially the liver. IU’s supercomputing resources were then used to simulate the process in thousands of virtual patients over the course of several days – an experiment that would have otherwise required several years to complete.
“These three submodels can also be run independently, which greatly simplifies the calibration of the model as well as the ability to connect the model’s predictions to clinically accessible measurements from real patients,” Sluka said. “So we can directly connect measurements from the clinic or the lab to the measurements predicted by our mathematical and computational models.”
Moreover, Sluka pointed out that the team’s virtual liver model was built using open source software, including technology developed by the Biocomplexity Institute called CompuCell3D, so other researchers can reuse all or part of the software without restriction; modify it to examine the toxic risk of different drugs or environmental toxins; or use it to study the toxicity of these compounds in nonhuman species.
More information: EurekAlert!

Comments are closed, but trackbacks and pingbacks are open.